Tutorial: Using the RIME Web Server
Overview
Welcome to the RIME Web Service. This service allows users to predict RNA-RNA interactions solely from sequence information. Starting from two long RNA sequences, the application calculates RIMEfull prediction scores for 200x200 nucleotides windows with a 100 nucleotides step. This tutorial will guide you through the steps of using the web service, from inputting your data to retrieving your results
Step-by-Step Instructions
Step 1: Accessing the Web Server
- Open your web browser and navigate to the https://tools.tartaglialab.com/rna_rna
Step 2: Inputting Your Data
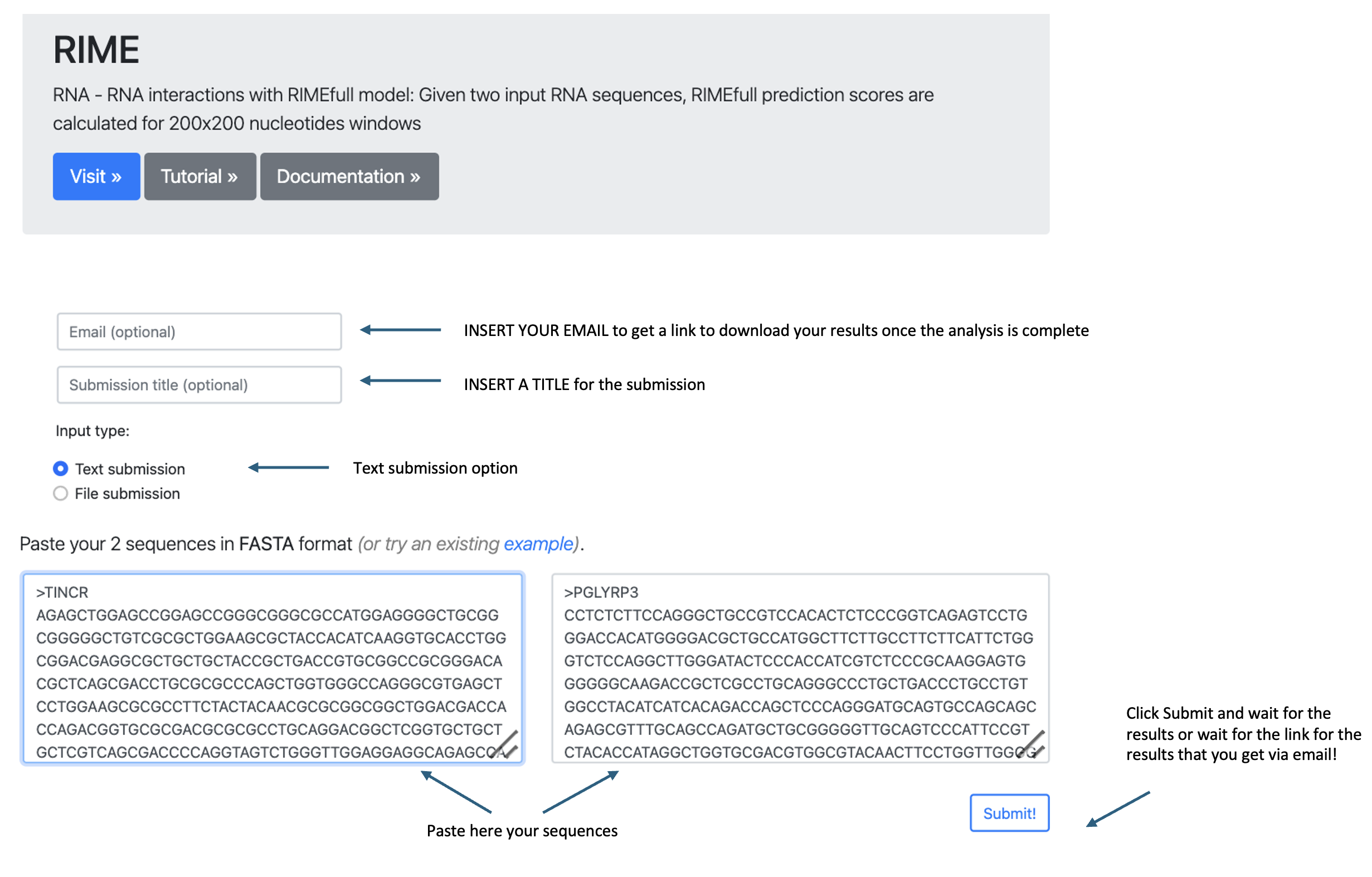
You can input your data in one of the following ways:
- Text submission:
- Scroll down to the input window labeled "Text submission".
- Type or paste your two RNA sequences in the FASTA format.
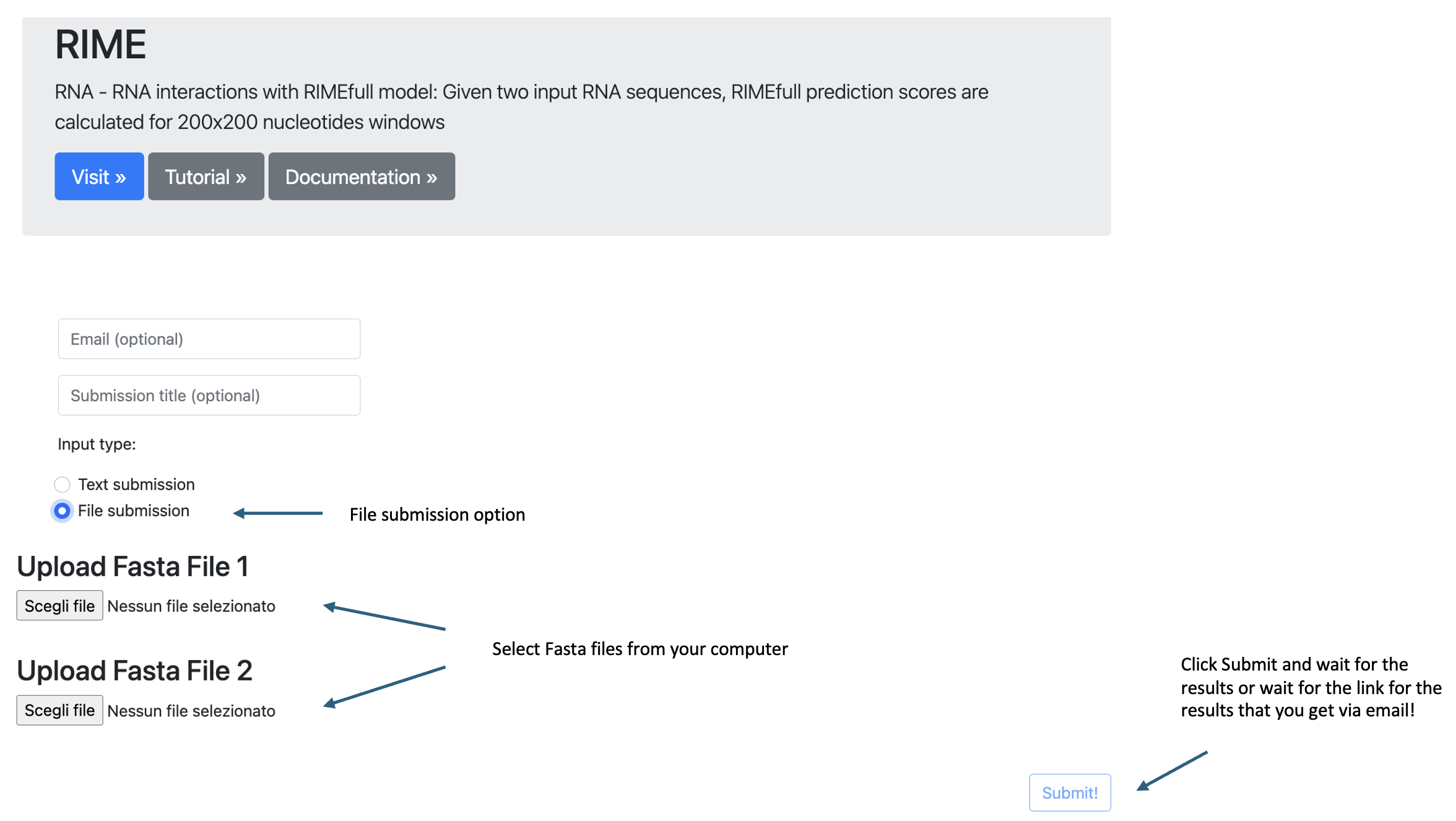
- File submission:
- Click on the "File submission" button.
- Select 2 files from your computer. Each file should be a FASTA.
Step 3:Submitting Your Data
- Submit:
- Click the "Submit" button to start the analysis process.
- You will be redirected to a page that confirms your submission and provides a tracking ID (web link).
Step 4: Retrieving Your Results
- Notification:
- If you provided an email address, you will receive an email with a link to your results once the analysis is complete
- Download Results:
- Click on the provided link to access your results.
- Table View: Navigate to the Table tab to view a detailed table displaying RIMEfull prediction scores across all 200x200 nucleotide windows. The same table can be downloaded in BEDPE format by clicking on the “Get table” button.
- Plot View: Switch to the Plot tab to visualize the same data as a heatmap for an intuitive representation of the results.
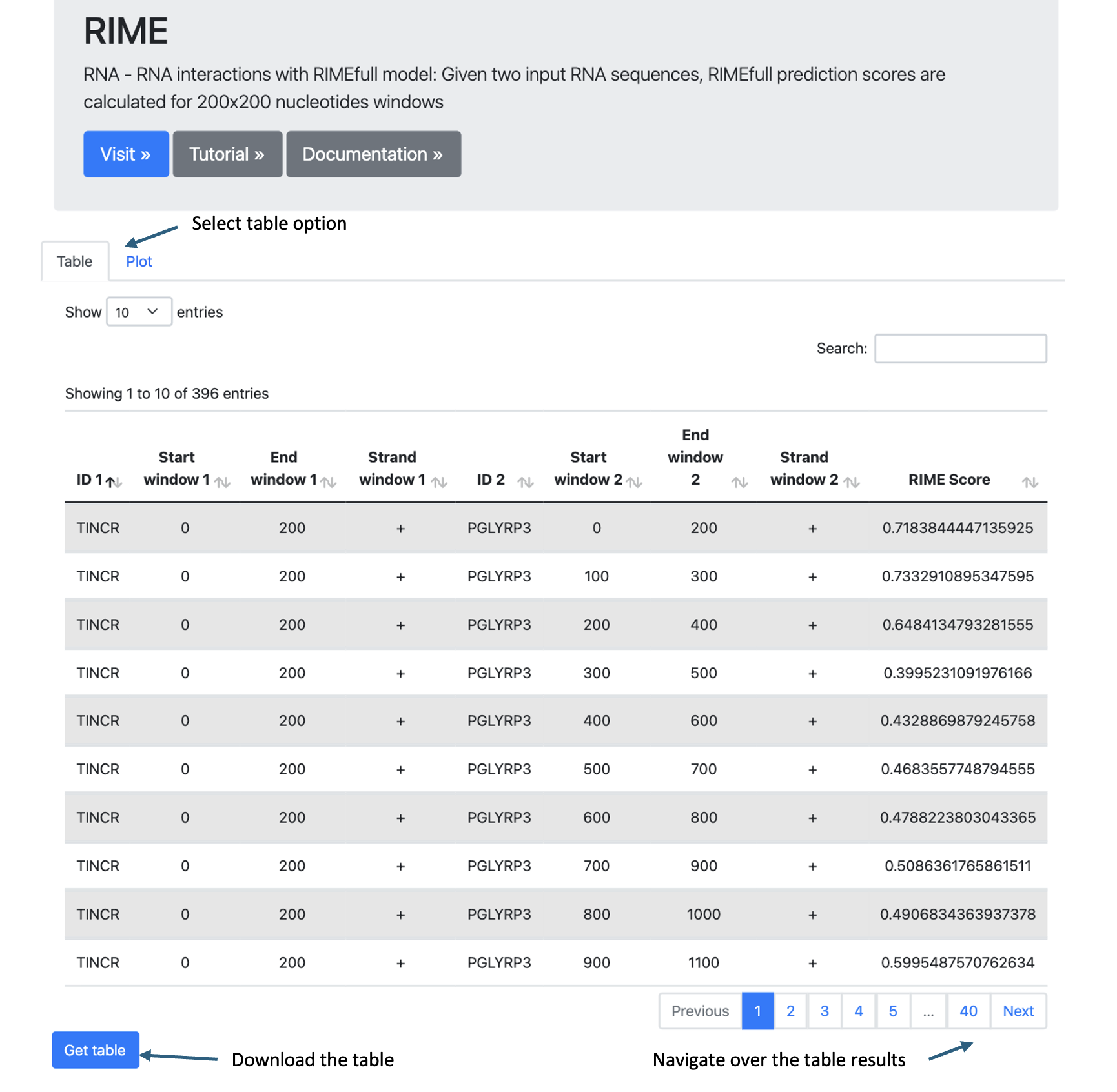
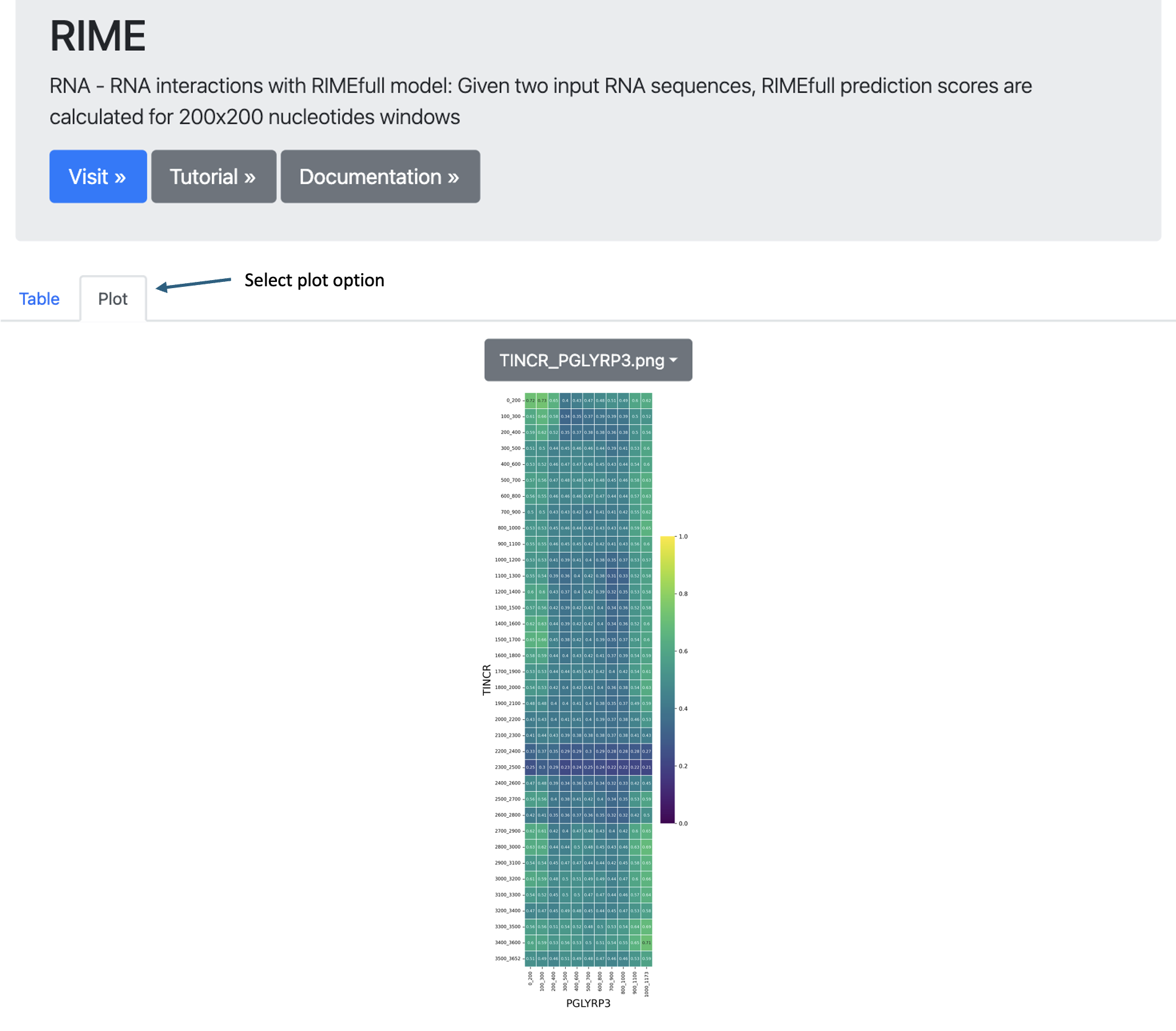
Table Description
This table represents pairwise RNA fragment interactions, where each row corresponds to a pairwise combination between two RNA sequence fragments and their interaction score. The coordinate system is zero-based, half-open, meaning that the coordinate of the first nucleotide of the sequence is 0 and that the end coordinate is 1 + the actual end position.
Plot Description: RNA Interaction Heatmap
This plot is a heatmap representation of the contact matrix between two RNA sequences, where each cell corresponds to an interaction score between a specific fragment of the first RNA and a specific fragment of the second RNA.
This visualization helps identify regions of strong interaction between the two RNA sequences, facilitating the study of potential RNA-RNA binding sites and functional interactions.
Additional Information
- Analysis Time:
- RIME typically completes within a few minutes for most pairs.
- Troubleshooting:
- File Upload Issues:
- Ensure your files are in the correct FASTA format. Both T and U characters are allowed to represent uracil nucleotides.
- No Results Email:
- Check your spam/junk folder if you do not receive an email within the expected timeframe.
- Ensure you entered the correct email address.