Tutorial: Using the catGRANULE 2.0 ROBOT Web Server
Overview
Welcome to the catGRANULE 2.0 ROBOT Server. This service allows users to analyze the LLPS propensity of proteins and perform mutagenesis analysis on individual protein sequences. This tutorial will guide you through the steps of using the web server, from inputting your data to retrieving your results.
Step-by-Step Instructions
Step 1: Accessing the Web Server
- Open your web browser and navigate to the https://tools.tartaglialab.com/
Step 2: Inputting Your Data
You can input your data in one of the following ways:
Option A: Upload a File- Choose a File:
- Click on the "Choose a file" button
- Select a file from your computer. The file can be:
- A FASTA file (.fasta)
- A PDB file (.pdb)
- Enter a Sequence:
- Scroll down to the input window labeled "Enter a sequence in FASTA format".
- Type or paste your protein sequence in the FASTA format.
Step 3: Selecting Analysis Options
Option A: LLPS Propensity Analysis- Default Option:
- By default, the server will perform LLPS propensity analysis on the inputted protein(s).
- Enable Mutagenesis Analysis:
- If you have uploaded a single protein file (either FASTA or PDB), you can opt for mutagenesis analysis.
- Check the box labeled "Perform mutagenesis analysis".
Step 4: Optional: Enter Your Email Address
- Highly Recommended:
- Enter your email address in the provided field. This will ensure you receive a notification with a link to download your results once the analysis is complete.
Step 5: Submitting Your Data
- Submit:
- Click the "Submit" button to start the analysis process.
- You will be redirected to a page that confirms your submission and provides a tracking ID (web link).
Step 6: Retrieving Your Results
- Notification:
- If you provided an email address, you will receive an email with a link to your results once the analysis is complete
- Download Results:
- Click on the provided link to access your results.
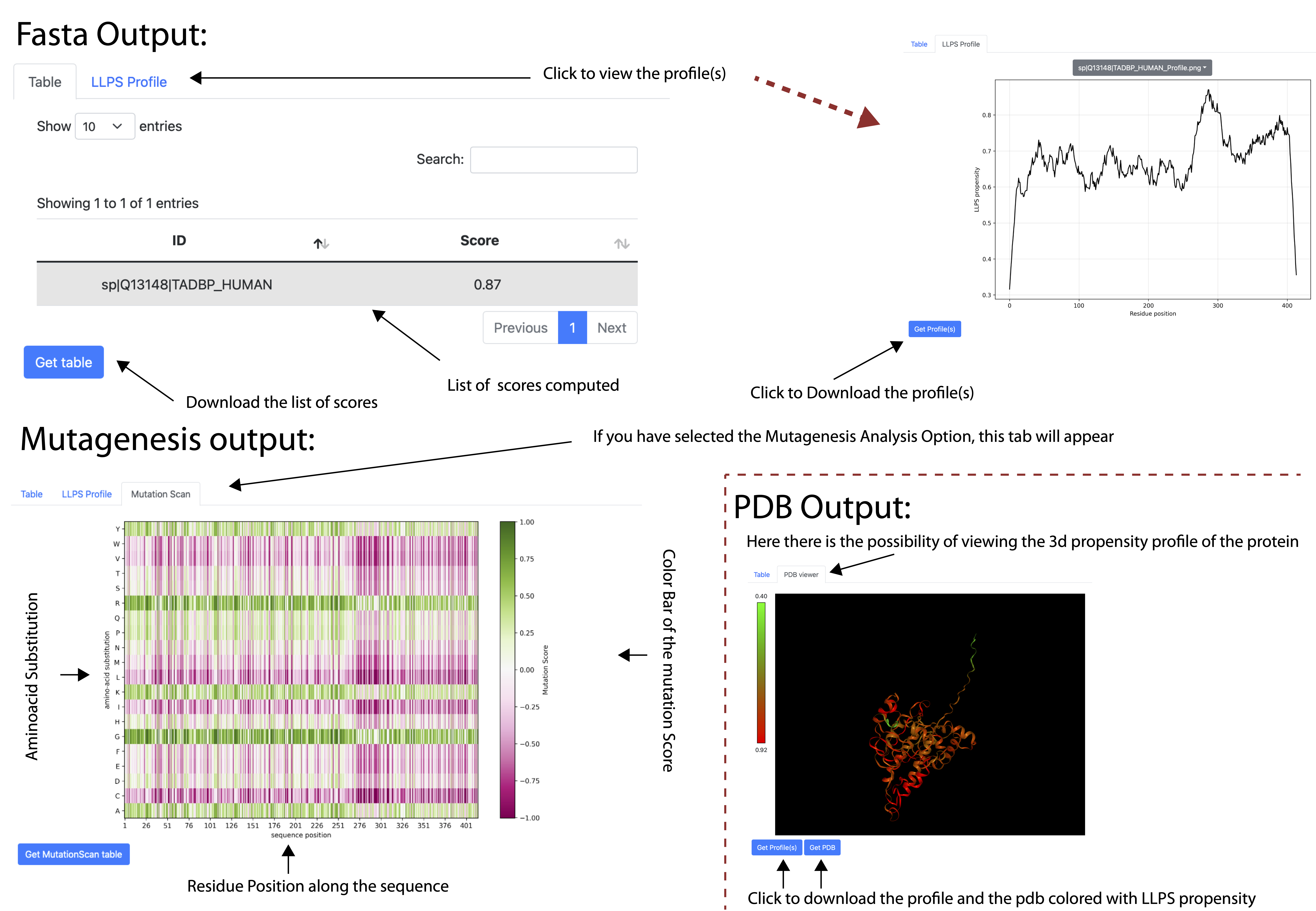
- The results page will display the LLPS propensity scores for each protein, moreover you get the LLPS propensity profile for each protein in separated txt (you can get the image in a png file).
- If you opted for mutagenesis analysis, a detailed report of the mutation scores for all possible amino acid substitutions will be included. Including the heat map of the mutation score per aminoacid and position.
Additional Information
- File Formats:
- FASTA: Plain text file format for representing protein sequences.
- PDB: Protein Data Bank file format, used for 3D structures of proteins. You can use the alphafold database to take the PDB files precomputed by the AI: https://alphafold.ebi.ac.uk/
- Analysis Time:
- LLPS propensity analysis typically completes within a few minutes for most files. For instance for a fasta submission we estimate: 1 seconds for one protein of 100 aa, 2 minutes for 100 proteins of around 100 aa. For a PDB submission the analysis needs around 1 minute.
- Mutagenesis analysis may take significantly longer depending on the length and complexity of the protein sequence. The estimation time is of 5 minutes for 100 aa protein, but it increases abruptly for larger protein, given the intrinsic exponential number of additional computations.
Troubleshooting
- File Upload Issues:
- Ensure your files are in the correct format and do not exceed the maximum allowed size (check the website for limits).
- No Results Email:
- Check your spam/junk folder if you do not receive an email within the expected timeframe.
- Ensure you entered the correct email address.
- Error Messages:
- If you encounter an error message, refer to the provided instructions on the web server for specific troubleshooting steps or contact support.
Graphical Example
INPUT Page: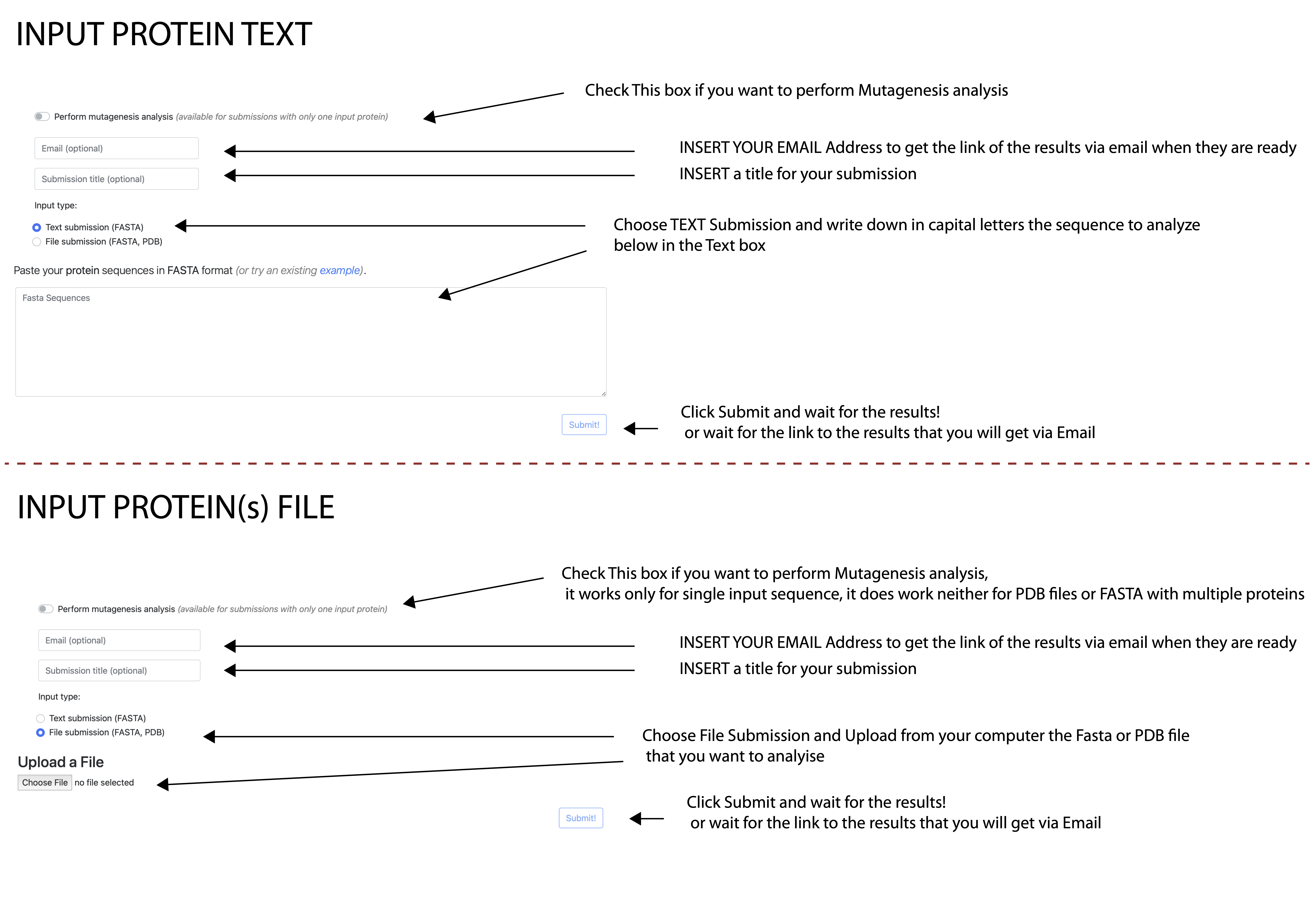 OUTPUT analysis:
OUTPUT analysis: